Small Samples and \(t\)-Tests#
What do we do when the overall sample size for the \(t\)-test is \(n < 40\)?
The process is the same as are the formulas, but we take a different approach to verifying the assumptions. For small sample sizes, we don’t have enough data points for the density plots and QQ plots to work properly. Thus, our normality check works like this:
Stem Plot – need evidence the sample was drawn from a bell-shaped distribution.
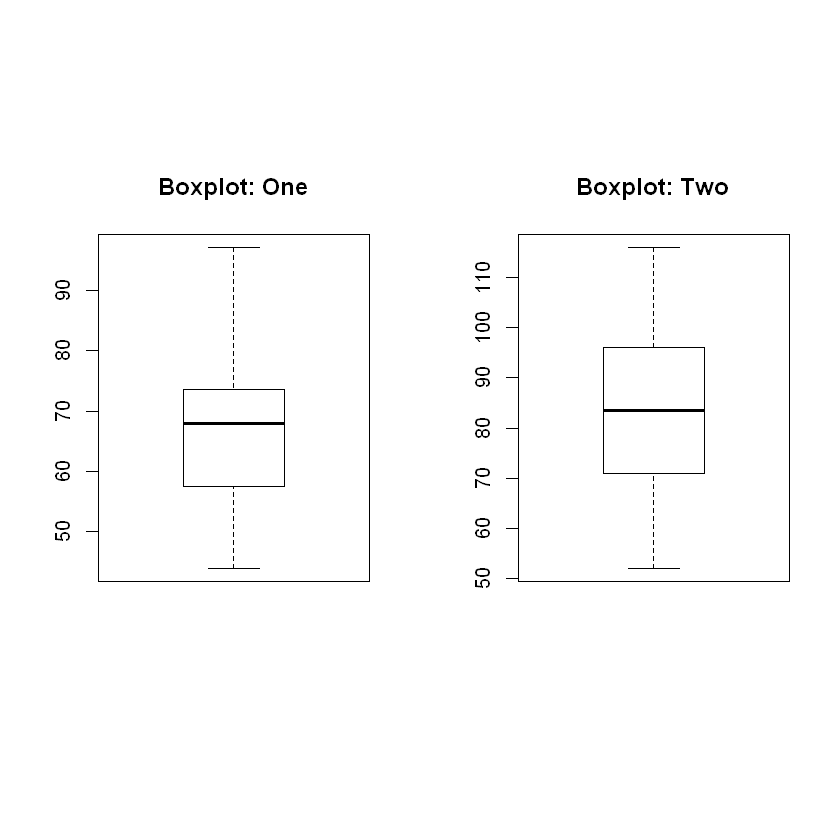
Box Plot – need to find zero outliers, as outliers are extremely influential in small samples.
For the homogeneity of the variances check, we still require the ratio of sample sizes to be no larger than \(2 : 1\).
Example: Independent Samples \(t\)-Test#
Adding bran to the diet can help patients with diverticulosis. The researchers wish to test two different preparations on patients. The transit time through the alimentary canal is tested for both groups. Which bran-based treatment works better? The data is shown below:
| Treat 1 | 44 | 51 | 52 | 55 | 60 | 62 | 66 | 68 | 69 | 71 | 71 | 76 | 82 | 91 | 97 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Treat 2 | 52 | 64 | 68 | 74 | 79 | 83 | 84 | 88 | 95 | 97 | 101 | 116 |