ANOVA#
Ron Fisher recognized the value of the \(t\)-test as created by William Sealy Gossett and developed the mathematical extension needed to use the same ideas with more than 2 samples.
Example: Coping Humor#
Let’s question whether, in the personality data set, the average coping humor (CHS) varies based upon Primary Humor Style (PHS). We will first load and then subset the data properly:
pers <- read.csv('https://faculty.ung.edu/rsinn/data/personality.csv')
humor <- pers[ , c('CHS','PHS')]
head(humor,5)
| CHS | PHS |
|---|---|
| 28 | SE |
| 29 | SE |
| 30 | AG |
| 27 | SE |
| 24 | AF |
Hypotheses#
The null hypothesis states that the four group means will all be approximately equal. The four primary humor styles are Affiliative, Aggressive, Self-Defeating and Self-Enhancing and will be abbreviate as af, ag, sd and se respectively. The alternative is usually included in words as shown:
Verifying the Assumptions#
We need to perform the following steps:
Create the linear model usinig the function lm().
Create a vector of residuals based upon the model.
Display a density plot of the residuals (checking for evidence of normality).
Display a QQ plot for the residuals (checking that all dots are reasonably close to the QQ line).
These steps are all performed in the single code below directly below.
mod = lm(CHS ~ PHS, data = humor)
res = residuals(mod)
layout(matrix(c(1,2), ncol = 2), lcm(8))
plot(density(res), main = 'Density Plot: Humor')
plot2 <- { qqnorm(mod$residual, main = 'QQ Plot: Humor')
qqline(mod$residual, col = 'red') }
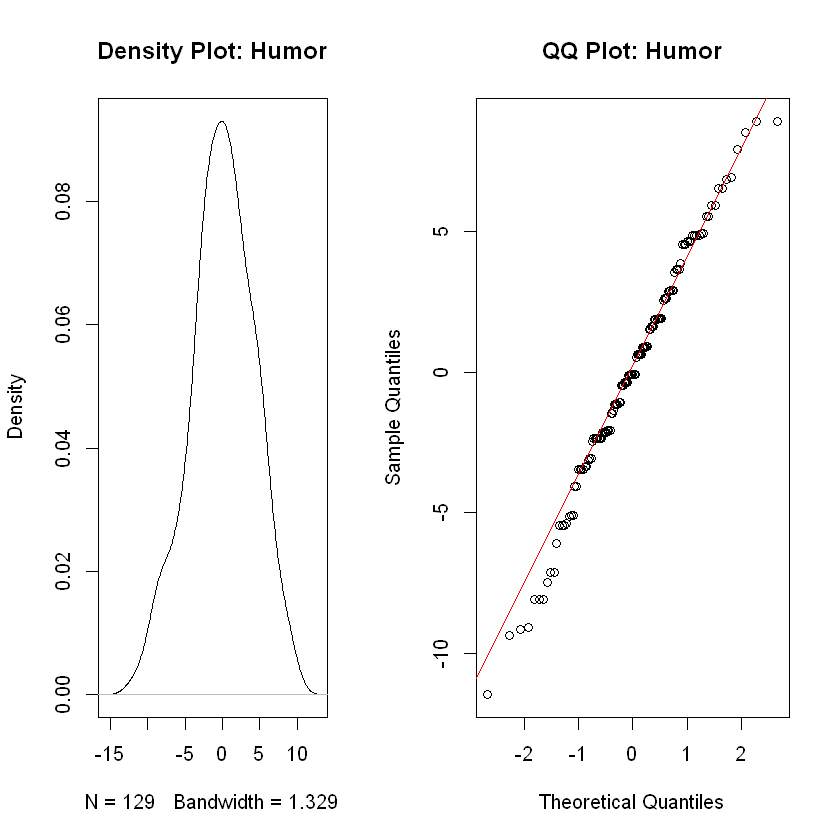
Analysis
The QQ plot is slightly concerning as the lower tail seems quite heavy, yet the density plot shows evidence these data were drawn from a normally distributed population. These data are appropriate for ANOVA methods though the heavy tail does indicate a small degree of concern about accuracy of \(p\)-values.
Conducting the ANOVA#
anova <- aov(mod)
summary(anova)
Df Sum Sq Mean Sq F value Pr(>F)
PHS 3 418.7 139.55 7.743 8.74e-05 ***
Residuals 125 2252.8 18.02
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Reporting Out#
With \(p = 8.74\times 10^{-5} < 0.05 = \alpha\), we reject the null. We have evidence, therefore, that at least one of the group means is significantly different than the others.
Tukey HSD Post Hoc Testing#
Whenever we reject the null in an ANOVA, we must conduct a post hoc test to ferret out all the significant differences between group means.
TukeyHSD(anova)
Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = mod)
$PHS
diff lwr upr p adj
AG-AF -1.385338 -4.1385491 1.3678724 0.5581859
SD-AF -2.100649 -4.9409691 0.7396704 0.2225615
SE-AF 2.669048 -0.2357284 5.5738237 0.0837472
SD-AG -0.715311 -3.3456923 1.9150703 0.8937175
SE-AG 4.054386 1.3545316 6.7542403 0.0008566
SE-SD 4.769697 1.9810664 7.5583275 0.0001080
The significant differences are indicated in rows 3, 5 and 6 where:
The upper and lower bounds of the confidence interval estimates both have the same sign, or
The indicated \(p\)-value is less than \(\alpha = 0.05\).
Thus, we have evidence for a difference between the means of the following pairs of groups:
Self-enhancing vs. Affiliative
Self-enhancing vs. Aggressive
Self-enhancing vs. Self-defeating

